Software at CIS : lddmm-volume : About
about | tutorial | user validation | remote validation | manual | namespace | faq | credits | changelog | feedback
Whole brain anatomical atlases at sub-millimeter resolution are emerging from the Human Brain Mapping and National Partnership in Advanced Computational Infrastructure (NPACI) initiatives. Analysis and inferences based on these anatomical volumes present a challenge in the emerging field of computational anatomy.
The Large Deformation Diffeomorphic Metric Mapping (LDDMM) tool is an application which aims to assign metric distances on the space of anatomical images in Computational Anatomy thereby allowing for the direct comparison and quantization of morphometric changes in shapes. As part of these efforts the Center for Imaging Science at Johns Hopkins University develop techniques to not only compare images, but also to visualize the changes and differences. This section will provide a brief overview of some of the applications of LDDMM and present the general idea behind the principles used.
LDDMM allows the user to calculate the metric distance between a given template image and a target image. The immediate implication of the tool is for uses in studying mental disorders such as Schizophrenia, Epilepsy, Alzheimer's and depression which have a structural manifestation on different parts of the brain including the Hippocampus and Basal Ganglia. It would be very valuable, for example, for a doctor to study the shape changes in the Basal Ganglia of a patient who has been experiencing Huntington's disease and act accordingly. The following image demonstrates the process of image matching and metric mapping on a Hippocampus.
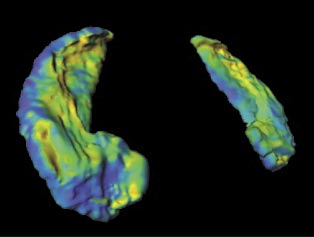
Another possible use of the tool would be in image segmentation. Image segmentation involves labelling different parts of the given image. In medical imaging
this is often required and precision is very important. The following graphic shows how LDDMM given a segmented template can produce the segmented target.
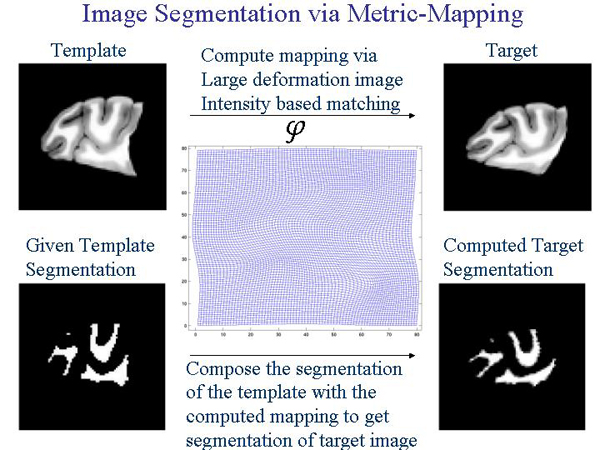
LDDMM in more technical terms gives the user a computational framework to study shape and size via metrics on flows of diffeomorphisms in a computational anatomy environment. The metric distances calculated are geodesic. LDDMM uses MPI and the SPMD architecture.
Last Modified: Thursday, 30th May, 2013 @ 09:26am






